Hybridisation
Hybridisation is the mating and production of offspring between individuals from morphologically and/or genetically distinct populations. The production of fertile (or semi-fertile) offspring allows alleles to pass from one population to another and will both increase genetic diversity within populations and reduce genetic distinctiveness among populations (Slatkin 1987; Harrison 1993).
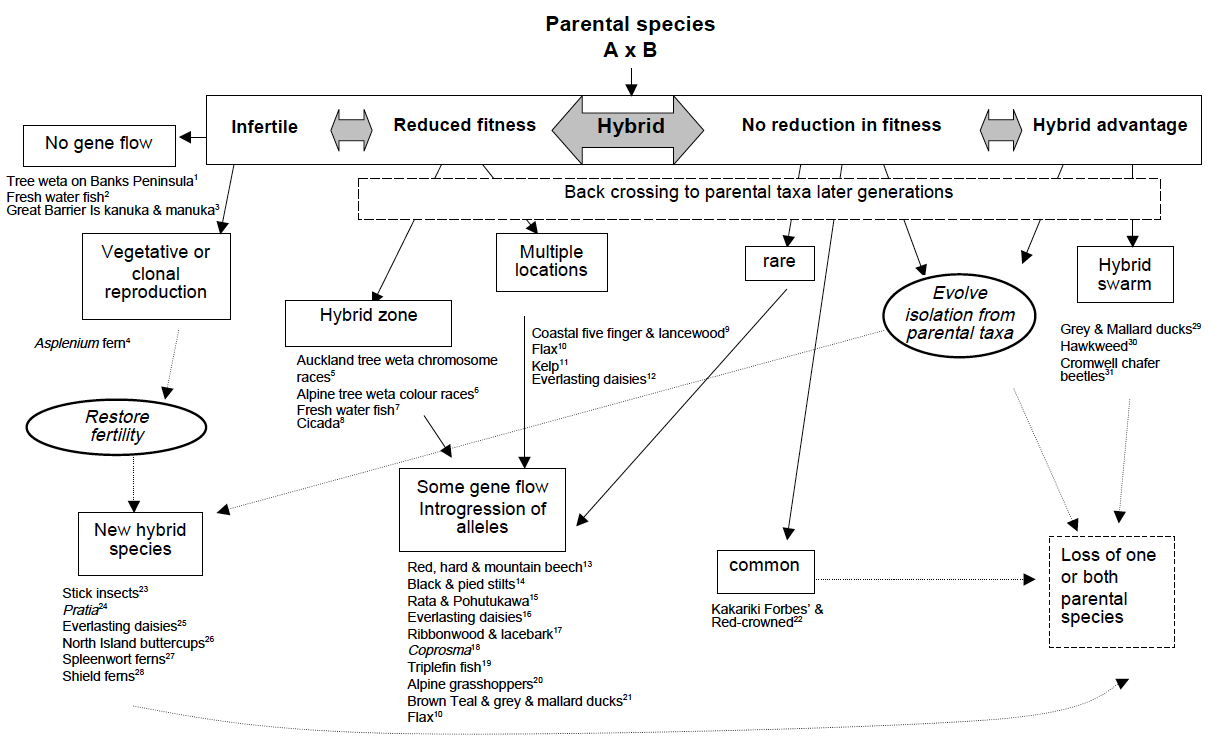
In New Zealand we have documented many examples of hybridisation (Morgan-Richards et al. 2009). Natural hybridisation can involve distinct species producing fertile hybrids (such as grasshoppers at Alexandra), and some examples of hybridisation involve morphologically distinct populations that are considered part of the same species (such as little and large geckos on Wellington’s south coast), still other examples of hybridisation involve individuals that are morphologically identical but genetically distinct, such as the chromosome races of the tree weta. New Zealand examples of natural hybridisation are reviewed in Morgan-Richards et al. 2009.
Members of the Phoenix lab study hybridisation because it provides a window into the understanding of evolutionary process such as speciation, selection, gene flow and the evolution of reproductive isolation.
Hybrid zones
Geckos
 Hybrid zones form where frequent interbreeding at the boundaries of distinct populations result in a region with a high frequency of offspring of mixed-population ancestry. Stable hybrid zones are maintained by a balance between dispersal of individuals and selection against hybrids. Where selection is dependent on the environment the distinct populations are frequently ecologically (and morphologically) differentiated. Sampling of a coastal transect of New Zealand geckos (Woodworthia maculata) was used to examine contact of morphologically differentiated populations. The gecko hybrid zone is unusual in the extent of size variation among individuals from either end of the hybrid zone. The larger geckos are twice the size of the small adult geckos, just 15 km away. Although the hybrid zone is narrow no evidence of assortative mating or hybrid disadvantage was found. Concordance of clines for adult gecko size and mitochondrial DNA was observed, as expected of a secondary contact, however the nuclear locus (RAG-2) had a much wider cline.
Hybrid zones form where frequent interbreeding at the boundaries of distinct populations result in a region with a high frequency of offspring of mixed-population ancestry. Stable hybrid zones are maintained by a balance between dispersal of individuals and selection against hybrids. Where selection is dependent on the environment the distinct populations are frequently ecologically (and morphologically) differentiated. Sampling of a coastal transect of New Zealand geckos (Woodworthia maculata) was used to examine contact of morphologically differentiated populations. The gecko hybrid zone is unusual in the extent of size variation among individuals from either end of the hybrid zone. The larger geckos are twice the size of the small adult geckos, just 15 km away. Although the hybrid zone is narrow no evidence of assortative mating or hybrid disadvantage was found. Concordance of clines for adult gecko size and mitochondrial DNA was observed, as expected of a secondary contact, however the nuclear locus (RAG-2) had a much wider cline.
Tree weta
 Tension zones are maintained by the interaction between selection against hybrids and dispersal of individuals. Investigating multiple hybrid zones within a single species provides the opportunity to examine differences in zone structure on a background of differences in extrinsic factors (e.g. age of the zone, ecology) or intrinsic factors (e.g. chromosomes). The New Zealand tree weta Hemideina thoracica comprises at least eight distinct chromosomal races with diploid numbers ranging from 2n 11 (XO) to 2n 23 (XO). Five independent hybrid zones were located that involve races differing from one another by a variety of chromosomal rearrangements. The predicted negative correlation between extent of karyotypic differentiation (measured in terms of both percent of genome and number of rearrangements) and zone width was not found. Conversely, the widest zones were those characterized by two chromosome rearrangements involving up to 35% of the genome. The narrowest zone occurred where the two races differ by a single chromosome rearrangement involving approximately 2% of the genome. The five estimates of chromosomal cline width ranged from 0. 5 km to 47 km. A comparative investigation of cline width for both chromosomal and mitochondrial markers revealed a complex pattern of zone characteristics. Three of the five zones showed cline concordance for the nuclear and cytoplasmic markers, and at two of the zones the clines were also coincident. Zones with the widest chromosomal clines had the widest mitochondrial DNA clines. It appears that, even within a single species, the extent of karyotypic differentiation between pairs of races is not a good predictor of the level of disadvantage suffered by hybrids. (Morgan-Richards & Wallis 2003)
Tension zones are maintained by the interaction between selection against hybrids and dispersal of individuals. Investigating multiple hybrid zones within a single species provides the opportunity to examine differences in zone structure on a background of differences in extrinsic factors (e.g. age of the zone, ecology) or intrinsic factors (e.g. chromosomes). The New Zealand tree weta Hemideina thoracica comprises at least eight distinct chromosomal races with diploid numbers ranging from 2n 11 (XO) to 2n 23 (XO). Five independent hybrid zones were located that involve races differing from one another by a variety of chromosomal rearrangements. The predicted negative correlation between extent of karyotypic differentiation (measured in terms of both percent of genome and number of rearrangements) and zone width was not found. Conversely, the widest zones were those characterized by two chromosome rearrangements involving up to 35% of the genome. The narrowest zone occurred where the two races differ by a single chromosome rearrangement involving approximately 2% of the genome. The five estimates of chromosomal cline width ranged from 0. 5 km to 47 km. A comparative investigation of cline width for both chromosomal and mitochondrial markers revealed a complex pattern of zone characteristics. Three of the five zones showed cline concordance for the nuclear and cytoplasmic markers, and at two of the zones the clines were also coincident. Zones with the widest chromosomal clines had the widest mitochondrial DNA clines. It appears that, even within a single species, the extent of karyotypic differentiation between pairs of races is not a good predictor of the level of disadvantage suffered by hybrids. (Morgan-Richards & Wallis 2003)
Stick insects
Hybridisation can result in new species. The stick insect genus Acanthoxyla probably arose from a hybrid between two distinct species (Morgan-Richards & Trewick 2005). All species of Acanthoxyla are all-female, and individuals reproduce without males (parthenogenetic reproduction). All species of Acanthoxyla feed on a range of native and introduced plant species. They vary in colour (green, brown, grey, speckled) and in how spiny they are (some individuals are covered in black-tipped spines from head to abdomen, some are smooth), and some Acanthoxyla are diploid and some triploid (Myers et al. 2012).
Biogeography 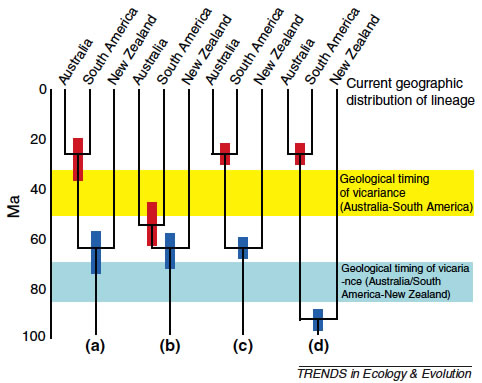
Biogeography explores the way species are distributed around the earth. It is in part descriptive (what species or combinations of species where), but increasingly has a more robust hypothesis testing objective. A valuable development in recent years has been in the inclusion of molecular phylogenetic information which can tell us about the relatedness of species in different areas of the globe, and also provide estimates of how far back in the past species shared common ancestors. It cannot, although this is often overlooked, tell us where in the world those ancestors existed. A particular problem in this respect is the effect on interpretation of the geographic history of plants and animals, of missing representatives of a group at a particular location. This might be be due to lack of sampling by researchers, extinction of a lineage at one or other place (or, worse everywhere!) or a true historical absence of a lineage at a place.
(See Crisp et al. 2011. Hypothesis testing in biogeography. Trends in Ecology and Evolution 1317: 1-7.)
Phylogeography
Phylogeography combines species’ geneologies with geographic distributions to study evolution.
New Zealand has long been a conundrum to biogeographers, possessing as it does geophysical and biotic features characteristic of both an island and a continent. This schism is reflected in provocative debate among dispersalist, vicariance biogeographic and panbiogeographic schools. A strong history in biogeography has spawned many hypotheses, which have begun to be addressed by a flood of molecular analyses. The time is now ripe to synthesize these findings on a background of geological and ecological knowledge. It has become increasingly apparent that most of the biota of New Zealand has links with other southern lands (particularly Australia) that are much more recent than the breakup of Gondwana. A compilation of molecular phylogenetic analyses of ca 100 plant and animal groups reveals that only 10% of these are even plausibly of archaic origin dating to the vicariant splitting of Zealandia from Gondwana. Effects of lineage extinction and lack of good calibrations in many cases strongly suggest that the actual proportion is even lower, in keeping with extensive Oligocene inundation of Zealandia. A wide compilation of papers covering phylogeographic structuring of terrestrial, freshwater and marine species shows some patterns emerging. These include: east–west splits across the Southern Alps, east– west splits across North Island, north–south splits across South Island, star phylogenies of southern mountain isolates, spread from northern, central and southern areas of high endemism, and recent recolonization (postvolcanic and anthropogenic). Excepting the last of these, most of these patterns seem to date to late Pliocene, coinciding with the rapid uplift of the Southern Alps. The diversity of New Zealand geological processes (sinking, uplift, tilting, sea level change, erosion, volcanism, glaciation) has produced numerous patterns, making generalizations difficult. Many species maintain pre-Pleistocene lineages, with phylogeographic structuring more similar to the Mediterranean region than northern Europe. This structure reflects the fact that glaciation was far from ubiquitous, despite the topography. Intriguingly, then, origins of the flora and fauna are island-like, whereas phylogeographic structure often reflects continental geological processes.
(Wallis & Trewick 2009. New Zealand phylogeography: evolution on a small continent. Molecular Ecology, 18, 3548–3580., Trewick et al. 2011 The invertebrate life of New Zealand: a phylogeographic approach. Insects. http://www.mdpi.com/journal/insects/ )
Assembly of the New Zealand fauna
The avifauna of New Zealand is taxonomically and ecologically distinctive, as is typical of island biotas. Molecular phylogenetics has revealed that 'tramp' species (widely dispersing taxa) have arrived in New Zealand even in the last few hundred years, and that some avian taxa have close phylogenetic relatives overseas (predominantly Australian), indicating their recent ancestors were tramps, too. Distinctive taxa with deep phylogenetic ancestry might be 'vicars' that owe their presence to vicariance, but lack of close morphological, taxonomic and phylogenetic affinity provides only tenuous evidence for this. Disproving the alternative possibility that apparent vicars are descended from tramps that dispersed in earlier times remains challenging, but molecular analyses have yielded startling insights. Among New Zealand's iconic taxa, the world's largest eagle shared a Pleistocene ancestor with a small Australian eagle, and giant, flightless moa are phylogenetic sisters of the much smaller, flying tinamous of South America. The New Zealand avifauna is neither isolated nor stable, but demonstrative of prolonged and ongoing colonization, speciation and extinction.
See Goldberg et al. 2008 Evolution of New Zealand's terrestrial fauna: a review of molecular evidence. Philosophical Transactions of the Royal Society, London 363: 3319–3334
Trewick & Gibb 2010. Vicars, tramps and assembly of the New Zealand avifauna: a review of molecular phylogenetic evidence. IBIS. 152: 226-253. Trewick 2011. Vicars & Vagrants. Ausralian Science.
Long distance dispersal
It is well established that many groups of plants and animals have undergone long-distance dispersal, but the extent to which this continues beyond initial colonization is largely unknown. To provide further insight into the frequency of gene flow mediated by long-distance dispersal, we investigated the origins of the fern Asplenium hookerianum on the Chatham Islands by comparing chloroplast trnL–trnF DNA sequence data from Chatham Islands’ A. hookerianum with extensive phylogeographic data for this
genetically variable species in mainland New Zealand. Our sequencing revealed two haplotypes in Chatham Islands’ A. hookerianum. These two haplotypes were each more closely related to haplotypes found in New Zealand than to each other. Despite the rarity of A. hookerianum on the Chatham Islands, its populations there appear to derive from at least two long-distance dispersal events from New Zealand, these possibly originating from different areas. We suggest that long-distance transoceanic dispersal, and the gene flow it can mediate, may be more common than is generally appreciated.
See Shepherd et al. 2009. Multiple colonizations of a remote oceanic archipelago by one species: how common is long-distance dispersal? Journal of Biogeography 36: 1972-1977.
Geographic parthenogenesis
Worldwide, parthenogenetic reproduction has evolved many times in the stick insects (Phasmatidae). Many parthenogenetic stick insects show the distribution pattern known as geographic parthenogenesis, in that they occupy habitats that are at higher altitude or latitude compared with their sexual relatives. Although it is often assumed that, in the short term, parthenogenetic populations will have a reproductive advantage over sexual populations; this is not necessarily the case. In New Zealand the stick insect Clitarchus hookeri has both sexual and asexual populations. Males are common in the northern half of the species’ range but rare or absent elsewhere, and we found that most C. hookeri from putative-parthenogenetic populations share a common ancestor. Female stick insects from bisexual populations of Clitarchus hookeri are capable of parthenogenetic reproduction, but those insects from putative-parthenogenetic populations produced few offspring via sexual reproduction when males were available. We found similar fertility (hatching success) in mated and virgin females. Mated females produce equal numbers of male and female offspring. Eggs from unmated females took longer to hatch, and all offspring were female. It appears that all C. hookeri females are capable of parthenogenetic reproduction, and thus could benefit from the numerical advantage this yields. Nevertheless, our phylogeographic evidence shows that the majority of all-female populations over a wide geographic area originate from a single loss of sexual reproduction.
See Morgan-Richards et al. 2010. Geographic parthenogenesis and the common tea-tree stick insect of New Zealand. Molecular Ecology 19(6): 1227-1238.
New Zealand Ecology and Ecophysiology
The distribution and abundance of species can be investigated by focusing on biotic and abiotic interactions. We are interested in combining phylogeographic patterns that inform us about the historic distribution of species, with ecological data about what limits species ranges’. Species ranges’ can be limited by environmental factors such as climate, and competition and predation, and resources such as pollinators and food sources. These environmental factors interact with historical climate change and population adaptation.
Comparative evolutionary ecology of tree weta Hemideina crassidens and H. thoracica.
Although two species of tree weta are common and abundant in North Island New Zealand, their ranges rarely overlap. Isolated populations of Hemideina crassidens in central North Island are entirely surrounded by H. thoracica. Although H. crassidens thrives at sea level, it appears that it is excluded by competition from H. thoracica, expect at high altitude and in frost-flats. The physiological basis to competitive exclusion of tree weta (Hemideina crassidens and H. thoracica) is being explored using measurements of resting metabolic rates and differing temperaturers.
What Limits a Weta?
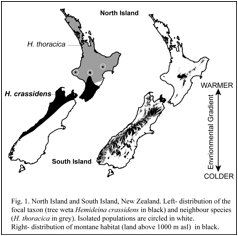
The primary goal of our research is to test the theory that gene flow limits species’ ranges by preventing adaptation. J.B.S. Haldane (1956) proposed that adaptation at the periphery of a species’ range might be prevented by gene flow from the centre of the species’ range where individuals are more numerous. Theoretical work since the mid 1990’s confirms that the flow of alleles (individuals, seeds or gametes) from populations in optimal habitat (source populations) into populations in suboptimal habitat (sink populations) could prevent adaptation to marginal environments by genetic swamping and thus limit the species’ distribution (Lenormand 2002). This theory makes the general prediction that populations without gene flow will occupy more extreme habitats compared to populations with high gene flow. If gene flow limits adaptation to the extremes of mountain living, as predicted by theoretical models, then populations of the wētā Hemideina crassidens isolated in cold-environments will be better adapted than non-isolated populations living at similar altitudes.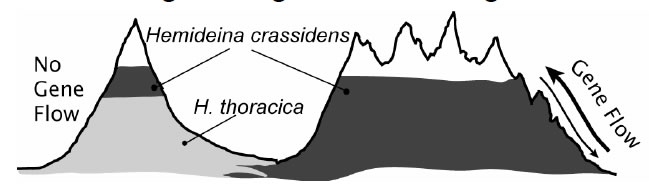 Furthermore, an isolated population will extend its range higher up the mountain into more extreme conditions. Alternatively, all populations at high altitude may be equally well adapted to their environment because the intensity of selection overwhelms gene flow or because gene flow is minimised by assortative mating (ecological speciation Schluter 2009). We can differentiate between these alternatives using population genetic tools.
Furthermore, an isolated population will extend its range higher up the mountain into more extreme conditions. Alternatively, all populations at high altitude may be equally well adapted to their environment because the intensity of selection overwhelms gene flow or because gene flow is minimised by assortative mating (ecological speciation Schluter 2009). We can differentiate between these alternatives using population genetic tools.
Biodiversity
Biodiversity can be measured at many levels, from ecosystem to genetics, but most often is examined at the level of species. Many distinct species means high biodiversity. On earth the majority of species level diversity is microscopic, each individual consisting of a single cell – bacteria, protozoa, amoeba, flagella, algae, protists, some fungi and archaea. However, when we think about conserving biodiversity we usually think about giant pandas, kakapo and whales. To ensure one large vertebrate species does not become extinct usually requires a whole ecosystem for that species to inhabit – and thus all the plants and invertebrates and unicellular organisms within the forest or ocean will also be preserved. But when we lose a rare habitat, or an unusual species we often have little knowledge of the dozens of species that use and require that space or resource. Thus one important aspect of conservation is to document biodiversity – knowing what we have before we loose it.

Within the Phoenix group we are carrying out many studies that inform about New Zealand’s biodiversity. Research projects such as investigating the systematics of New Zealand cave weta, and the NZ ground weta (Hemiandrus) lead directly to a better understanding of our biodiversity. These studies are naming new species and describing where they are found and their whakapapa. Our projects sometimes focus on endangered species to identify conservation units and highlight populations that need attention (Powellphanta; Trewick et al. 2008; Trewick 1999), and our genetic studies can lead to a better understanding of the dangers of population fragmentation (skinks), and where species boundaries are (gecko). On the East Coast on North Island we are documenting the numbers of species of marine snails living in rock pools and by using species ranges and genetic data can learn how the currents and coast line are connecting and separating populations. This information will link directly to iwi plans for a rahui (protected area). But studies of species interactions and ecosystems services are also important for understanding biodiversity and the way unrelated taxa rely on each other for food or pollination or seed dispersal. Parasitic wasps need aphids to eat, weta need trees to hide in and leaves to eat, orchids need gnats to transfer pollen, thus all these studies have biodiversity implications.
Conservation Genetics
Conservation biology makes use of genetic tools to both delimit species boundaries and understand and manage the genetic consequences of small populations.
Many population genetic and phylogenetic studies improve our understanding of the specific status of populations and thus are  important for management of endangered species, helping determine priorities and plan conservation programmes. In our lab we have been involved in the delimitation of species boundaries of critically endangered species such as the robust grasshopper (Brachaspis robustus) (Trewick 2001, Trewick & Morris 2008), giant land snails (Powelliphanta augusta) (Trewick et al. 2008, Walker et al. 2008) and geckos (Hoplodactylus duvaucilli), identifying cryptic species such as peripatus (Trewick 1998; 2000), and assessing conservation status of Philippine freshwater crocodile (Crocodylus mindorensis). Genetic studies sometimes reveal fewer species exist than the taxonomy suggests (e.g. stick insects (Trewick et al. 2005), cave weta (Cook et al. 2010).
important for management of endangered species, helping determine priorities and plan conservation programmes. In our lab we have been involved in the delimitation of species boundaries of critically endangered species such as the robust grasshopper (Brachaspis robustus) (Trewick 2001, Trewick & Morris 2008), giant land snails (Powelliphanta augusta) (Trewick et al. 2008, Walker et al. 2008) and geckos (Hoplodactylus duvaucilli), identifying cryptic species such as peripatus (Trewick 1998; 2000), and assessing conservation status of Philippine freshwater crocodile (Crocodylus mindorensis). Genetic studies sometimes reveal fewer species exist than the taxonomy suggests (e.g. stick insects (Trewick et al. 2005), cave weta (Cook et al. 2010).

Endangered species can suffer from inbreeding depression and loss of genetic diversity, resulting from small popualtions. In our lab, students have studied crocodile conservation genetics and skink conservation genetics with this in mind. Other current research includes threatened grasshoppers, the status of New Zealand falcon, and population genetics of weka and Auckland Island rails.
There are lots of opportunities for research in Conservation Genetics in our lab.
Rates, dates & extinction
Latitudinal biodiversity gradient
Species diversity is higher in the tropics (low latitude) than in temperate regions (high latitude) resulting in a latitudinal biodiversity gradient. The latitudinal biodiversity gradient must be generated by differential rates of speciation and/or extinction and/or immigration among regions, but the role of each of these processes is still unclear. Recent studies examining differences in rates of molecular evolution have inferred a direct link between rate of molecular evolution and rate of speciation, and postulated these as important drivers of the latitudinal biodiversity gradient. Our group is examining the factors that might be responsible for differences in rates of molecular evolution. Critical to this is the directionality of the relationship between speciation rates and rates of molecular evolution. We are gathering molecular genetic evidence to understand if variation in the rate of molecular evolution has a role in the latitudinal biodiversity gradient. Using weta and snails from New Caledonia and New Zealand we can compare rates, diversity and population size in related taxa at different latitudes.
Calibrations of molecular clocks with living fossils
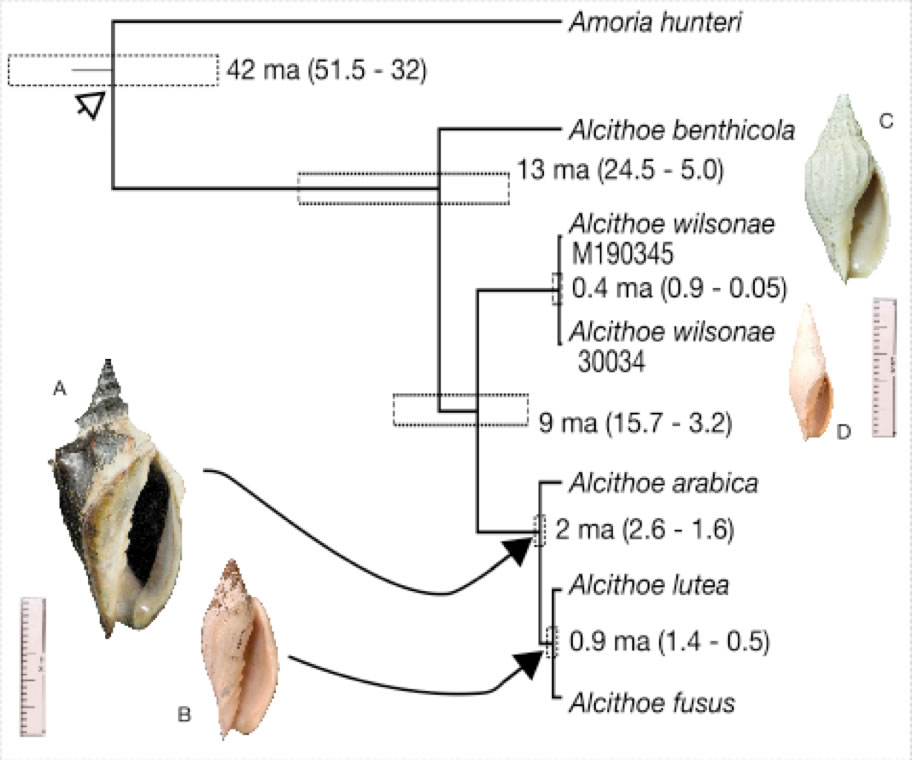 Species definition and delimitation is a non-trivial problem in evolutionary biology that is particularly problematic for fossil organisms. This is especially true when considering the continuity of past and present species, because species defined in the fossil record are not necessarily equivalent to species defined in the living fauna. Correctly assigned fossil species are critical for sensitive downstream analysis (e.g. diversification studies and molecular-clock calibration). The marine snail genus Alcithoe exemplifies many of the problems with species identification. The paucity of discriminatory characters, prevalence of morphological convergence between species and considerable variability within species, observed in Alcithoe are typical of a broad range of fossilised organisms. Using a synthesis of molecular and morphometric approaches we have shown that two taxa currently recognised as distinct are morphological variants of a single species. A concordance between the palaeontological record and divergence time of the lineage inferred using molecular-clock analysis has allowed us to validate the fossil record. This work demonstrates the utility of living species represented in the fossil record as candidates for molecular-clock calibration, as the veracity of fossil species assignment can be more rigorously tested.
Species definition and delimitation is a non-trivial problem in evolutionary biology that is particularly problematic for fossil organisms. This is especially true when considering the continuity of past and present species, because species defined in the fossil record are not necessarily equivalent to species defined in the living fauna. Correctly assigned fossil species are critical for sensitive downstream analysis (e.g. diversification studies and molecular-clock calibration). The marine snail genus Alcithoe exemplifies many of the problems with species identification. The paucity of discriminatory characters, prevalence of morphological convergence between species and considerable variability within species, observed in Alcithoe are typical of a broad range of fossilised organisms. Using a synthesis of molecular and morphometric approaches we have shown that two taxa currently recognised as distinct are morphological variants of a single species. A concordance between the palaeontological record and divergence time of the lineage inferred using molecular-clock analysis has allowed us to validate the fossil record. This work demonstrates the utility of living species represented in the fossil record as candidates for molecular-clock calibration, as the veracity of fossil species assignment can be more rigorously tested.
Punctuated Evolution
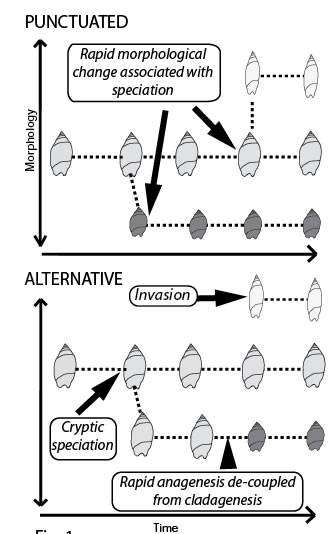 The study of fossils provides an impression of morphological evolution made up of long (boring) periods of constrained evolution when nothing changes interspersed with sudden leaps in shape. Many interpret these abrupt changes in morphology as being the result of speciation (punctuated equilibrium). We ask - Do geologically abrupt changes in fossil morphology result from speciation via cladogenesis? [Fig: Punctuated]. This question is central to debate surrounding punctuated equilibrium (see Eldredge & Gould 1972 Models in Paleobiology; Gould & Eldredge 1993. Nature 366: 223-227; Clarke 1994 Nature 368: 407; Levinton 1994 Nature 368: 407-408.). Using New Zealand marine snails with a superb fossil record (Crampton et al. 2003. Science 301: 358-360) and new DNA sequencing technology we plan to test rigorously the fundamental theory that cladogenesis is responsible for morphologically punctuated evolution.
The study of fossils provides an impression of morphological evolution made up of long (boring) periods of constrained evolution when nothing changes interspersed with sudden leaps in shape. Many interpret these abrupt changes in morphology as being the result of speciation (punctuated equilibrium). We ask - Do geologically abrupt changes in fossil morphology result from speciation via cladogenesis? [Fig: Punctuated]. This question is central to debate surrounding punctuated equilibrium (see Eldredge & Gould 1972 Models in Paleobiology; Gould & Eldredge 1993. Nature 366: 223-227; Clarke 1994 Nature 368: 407; Levinton 1994 Nature 368: 407-408.). Using New Zealand marine snails with a superb fossil record (Crampton et al. 2003. Science 301: 358-360) and new DNA sequencing technology we plan to test rigorously the fundamental theory that cladogenesis is responsible for morphologically punctuated evolution.
Molecular phylogenetics relies on the assumption that morphological punctuations are simultaneous with cladogenesis in order to calibrate molecular clocks using dated fossils. Although there is gathering evidence that fossil species in many invertebrates are likely to be biological species (based on detailed morphological and genetic work using extant taxa), other explanations are possible for the rapid morphological changes seen in the fossil record12,13 [Fig: Alternative]. For example ecotypes, hybridisation, cryptic species, punctuated anagenesis and biological invasion could all decouple geologically rapid morphological change from speciation.
We expect more direct evidence of the link between cladogenesis and morphological evolution will follow from studies of the phylogeography and paleontology of populations, species, and clades merged to produce a time-space integrated view of phenotypic divergence (Hunt 2010 - Evolution in fossil lineages: Paleontology and The Origin of Species. American Naturalist, 176: S61-S76.).
Species Interactions
Plant-insect interactions
The distinctiveness of New Zealand’s large endemic orthopterans (weta) and lack of small mammals in our forest ecosystems led to the description of weta as ecologically equivalent to rodents in other countries. However, if these taxa are to be compared, the details of their ecology are important and the scale of their influence in an ecosystem must be taken into account. In particular the ‘invertebrate mouse’ cliché is misleading. For example, reproductive potential and scale of change in population size differ greatly between mice (Mus musculus) and tree weta (Hemideina sp.). Endothermic mice have a metabolic rate almost 20 times faster than ectothermic tree weta, an intrinsic rate of increase some 275 times higher, and consume a high quality diet dominated by seeds and invertebrates and devoid of leaves, in contrast to tree weta diets. (See Griffin et al. 2011 Exploring the concept of niche convergence in a land without rodents: the case of weta as small mammals. New Zealand Journal of Ecology. In press.)
Mutualisms or interspecific interactions involving net mutual benefits, are an important component of ecological theory, although effectively demonstrating mutualism is notoriously difficult. Among two New Zealand endemics, a slightly elevated germination rate of Fuchsia excorticata seeds after passage through tree weta compared with seeds manually extracted from fruit, led to the proposal that a mutualistic relationship exists between this plant and animal. An improved germination rate, or any other single trait, however, does not alone constitute evidence for mutualism; the relative costs and benefits of numerous components of the interaction need to be accounted for. We considered the costs and benefits to F. excorticata of the putative seed dispersal mutualism with tree weta (Wyman et al. 2010). Tree weta provided with F. excorticata fruits destroyed 78% of the seeds they consumed, did not move fruit; and faeces containing seeds were deposited near their roost holes (which are naturally in trees). The seeds remaining after fruit consumption and those that are ingested but survive gut passage are unlikely to be deposited in suitable habitat for seedling survival. Plant food preferences of captive tree weta assessed using pairwise leaf choice tests showed that the leaves of F. excorticata were the least preferred of six commonly encountered plants. In addition, we found that tree weta did not show a preference for F. excorticata fruit over a standard leafy diet, indicating they are unlikely to be actively seeking fruit in preference to other sources of food. These observations indicate that any interaction between tree weta and F. excorticata is likely to be opportunistic rather than mutualistic, and highlight the difficulty of characterizing such interactions.
Both tree and ground wētā have been proposed as potential seed dispersers of some New Zealand fruit. We have examine evidence for coevolution of ground wētā and  fleshy fruits (Morgan-Richards, Trewick & Dunavan 2008). We found that although ground wētā consume fruits from Gaultheria depressa and G. antipoda, they do not do so in a way that would suggest they had coevolved as dispersers with these or other New Zealand plants. We observed a positive preference for eating fruits of plants with seeds that were too big for ground wētā to ingest. Thus there is little support to the proposal that ground wētā have coevolved with New Zealand plants resulting in the unusual characteristics displayed by many species (pale fruit presented within a divaricating canopy).
fleshy fruits (Morgan-Richards, Trewick & Dunavan 2008). We found that although ground wētā consume fruits from Gaultheria depressa and G. antipoda, they do not do so in a way that would suggest they had coevolved as dispersers with these or other New Zealand plants. We observed a positive preference for eating fruits of plants with seeds that were too big for ground wētā to ingest. Thus there is little support to the proposal that ground wētā have coevolved with New Zealand plants resulting in the unusual characteristics displayed by many species (pale fruit presented within a divaricating canopy).
Parasite-host interactions
 UNDER CONSTRUCTION
UNDER CONSTRUCTION



 Hybrid zones form where frequent interbreeding at the boundaries of distinct populations result in a region with a high frequency of offspring of mixed-population ancestry. Stable hybrid zones are maintained by a balance between dispersal of individuals and selection against hybrids. Where selection is dependent on the environment the distinct populations are frequently ecologically (and morphologically) differentiated. Sampling of a coastal transect of New Zealand geckos (Woodworthia maculata) was used to examine contact of morphologically differentiated populations. The gecko hybrid zone is unusual in the extent of size variation among individuals from either end of the hybrid zone. The larger geckos are twice the size of the small adult geckos, just 15 km away. Although the hybrid zone is narrow no evidence of assortative mating or hybrid disadvantage was found. Concordance of clines for adult gecko size and mitochondrial DNA was observed, as expected of a secondary contact, however the nuclear locus (RAG-2) had a much wider cline.
Hybrid zones form where frequent interbreeding at the boundaries of distinct populations result in a region with a high frequency of offspring of mixed-population ancestry. Stable hybrid zones are maintained by a balance between dispersal of individuals and selection against hybrids. Where selection is dependent on the environment the distinct populations are frequently ecologically (and morphologically) differentiated. Sampling of a coastal transect of New Zealand geckos (Woodworthia maculata) was used to examine contact of morphologically differentiated populations. The gecko hybrid zone is unusual in the extent of size variation among individuals from either end of the hybrid zone. The larger geckos are twice the size of the small adult geckos, just 15 km away. Although the hybrid zone is narrow no evidence of assortative mating or hybrid disadvantage was found. Concordance of clines for adult gecko size and mitochondrial DNA was observed, as expected of a secondary contact, however the nuclear locus (RAG-2) had a much wider cline.  Tension zones are maintained by the interaction between selection against hybrids and dispersal of individuals. Investigating multiple hybrid zones within a single species provides the opportunity to examine differences in zone structure on a background of differences in extrinsic factors (e.g. age of the zone, ecology) or intrinsic factors (e.g. chromosomes). The New Zealand tree weta Hemideina thoracica comprises at least eight distinct chromosomal races with diploid numbers ranging from 2n 11 (XO) to 2n 23 (XO). Five independent hybrid zones were located that involve races differing from one another by a variety of chromosomal rearrangements. The predicted negative correlation between extent of karyotypic differentiation (measured in terms of both percent of genome and number of rearrangements) and zone width was not found. Conversely, the widest zones were those characterized by two chromosome rearrangements involving up to 35% of the genome. The narrowest zone occurred where the two races differ by a single chromosome rearrangement involving approximately 2% of the genome. The five estimates of chromosomal cline width ranged from 0. 5 km to 47 km. A comparative investigation of cline width for both chromosomal and mitochondrial markers revealed a complex pattern of zone characteristics. Three of the five zones showed cline concordance for the nuclear and cytoplasmic markers, and at two of the zones the clines were also coincident. Zones with the widest chromosomal clines had the widest mitochondrial DNA clines. It appears that, even within a single species, the extent of karyotypic differentiation between pairs of races is not a good predictor of the level of disadvantage suffered by hybrids. (Morgan-Richards & Wallis 2003)
Tension zones are maintained by the interaction between selection against hybrids and dispersal of individuals. Investigating multiple hybrid zones within a single species provides the opportunity to examine differences in zone structure on a background of differences in extrinsic factors (e.g. age of the zone, ecology) or intrinsic factors (e.g. chromosomes). The New Zealand tree weta Hemideina thoracica comprises at least eight distinct chromosomal races with diploid numbers ranging from 2n 11 (XO) to 2n 23 (XO). Five independent hybrid zones were located that involve races differing from one another by a variety of chromosomal rearrangements. The predicted negative correlation between extent of karyotypic differentiation (measured in terms of both percent of genome and number of rearrangements) and zone width was not found. Conversely, the widest zones were those characterized by two chromosome rearrangements involving up to 35% of the genome. The narrowest zone occurred where the two races differ by a single chromosome rearrangement involving approximately 2% of the genome. The five estimates of chromosomal cline width ranged from 0. 5 km to 47 km. A comparative investigation of cline width for both chromosomal and mitochondrial markers revealed a complex pattern of zone characteristics. Three of the five zones showed cline concordance for the nuclear and cytoplasmic markers, and at two of the zones the clines were also coincident. Zones with the widest chromosomal clines had the widest mitochondrial DNA clines. It appears that, even within a single species, the extent of karyotypic differentiation between pairs of races is not a good predictor of the level of disadvantage suffered by hybrids. (Morgan-Richards & Wallis 2003) 


 Furthermore, an isolated population will extend its range higher up the mountain into more extreme conditions. Alternatively, all populations at high altitude may be equally well adapted to their environment because the intensity of selection overwhelms gene flow or because gene flow is minimised by assortative mating (ecological speciation Schluter 2009). We can differentiate between these alternatives using population genetic tools.
Furthermore, an isolated population will extend its range higher up the mountain into more extreme conditions. Alternatively, all populations at high altitude may be equally well adapted to their environment because the intensity of selection overwhelms gene flow or because gene flow is minimised by assortative mating (ecological speciation Schluter 2009). We can differentiate between these alternatives using population genetic tools. important for management of endangered species, helping determine priorities and plan conservation programmes. In our lab we have been involved in the delimitation of species boundaries of critically endangered species such as the robust grasshopper (Brachaspis robustus) (Trewick 2001, Trewick & Morris 2008), giant land snails (Powelliphanta augusta) (Trewick et al. 2008, Walker et al. 2008) and geckos (Hoplodactylus duvaucilli), identifying cryptic species such as peripatus (Trewick 1998; 2000), and assessing conservation status of Philippine freshwater crocodile (Crocodylus mindorensis). Genetic studies sometimes reveal fewer species exist than the taxonomy suggests (e.g. stick insects (Trewick et al. 2005), cave weta (Cook et al. 2010).
important for management of endangered species, helping determine priorities and plan conservation programmes. In our lab we have been involved in the delimitation of species boundaries of critically endangered species such as the robust grasshopper (Brachaspis robustus) (Trewick 2001, Trewick & Morris 2008), giant land snails (Powelliphanta augusta) (Trewick et al. 2008, Walker et al. 2008) and geckos (Hoplodactylus duvaucilli), identifying cryptic species such as peripatus (Trewick 1998; 2000), and assessing conservation status of Philippine freshwater crocodile (Crocodylus mindorensis). Genetic studies sometimes reveal fewer species exist than the taxonomy suggests (e.g. stick insects (Trewick et al. 2005), cave weta (Cook et al. 2010).

 Species definition and delimitation is a non-trivial problem in evolutionary biology that is particularly problematic for fossil organisms. This is especially true when considering the continuity of past and present species, because species defined in the fossil record are not necessarily equivalent to species defined in the living fauna. Correctly assigned fossil species are critical for sensitive downstream analysis (e.g. diversification studies and molecular-clock calibration). The marine snail genus Alcithoe exemplifies many of the problems with species identification. The paucity of discriminatory characters, prevalence of morphological convergence between species and considerable variability within species, observed in Alcithoe are typical of a broad range of fossilised organisms. Using a synthesis of molecular and morphometric approaches we have shown that two taxa currently recognised as distinct are morphological variants of a single species. A concordance between the palaeontological record and divergence time of the lineage inferred using molecular-clock analysis has allowed us to validate the fossil record. This work demonstrates the utility of living species represented in the fossil record as candidates for
Species definition and delimitation is a non-trivial problem in evolutionary biology that is particularly problematic for fossil organisms. This is especially true when considering the continuity of past and present species, because species defined in the fossil record are not necessarily equivalent to species defined in the living fauna. Correctly assigned fossil species are critical for sensitive downstream analysis (e.g. diversification studies and molecular-clock calibration). The marine snail genus Alcithoe exemplifies many of the problems with species identification. The paucity of discriminatory characters, prevalence of morphological convergence between species and considerable variability within species, observed in Alcithoe are typical of a broad range of fossilised organisms. Using a synthesis of molecular and morphometric approaches we have shown that two taxa currently recognised as distinct are morphological variants of a single species. A concordance between the palaeontological record and divergence time of the lineage inferred using molecular-clock analysis has allowed us to validate the fossil record. This work demonstrates the utility of living species represented in the fossil record as candidates for  The study of fossils provides an impression of morphological evolution made up of long (boring) periods of constrained evolution when nothing changes interspersed with sudden leaps in shape. Many interpret these abrupt changes in morphology as being the result of speciation (punctuated equilibrium). We ask - Do geologically abrupt changes in fossil morphology result from speciation via cladogenesis? [Fig: Punctuated]. This question is central to debate surrounding punctuated equilibrium (see Eldredge & Gould 1972 Models in Paleobiology; Gould & Eldredge 1993. Nature 366: 223-227; Clarke 1994 Nature 368: 407; Levinton 1994 Nature 368: 407-408.). Using New Zealand marine snails with a superb fossil record (Crampton et al. 2003. Science 301: 358-360) and new DNA sequencing technology we plan to test rigorously the fundamental theory that cladogenesis is responsible for morphologically punctuated evolution.
The study of fossils provides an impression of morphological evolution made up of long (boring) periods of constrained evolution when nothing changes interspersed with sudden leaps in shape. Many interpret these abrupt changes in morphology as being the result of speciation (punctuated equilibrium). We ask - Do geologically abrupt changes in fossil morphology result from speciation via cladogenesis? [Fig: Punctuated]. This question is central to debate surrounding punctuated equilibrium (see Eldredge & Gould 1972 Models in Paleobiology; Gould & Eldredge 1993. Nature 366: 223-227; Clarke 1994 Nature 368: 407; Levinton 1994 Nature 368: 407-408.). Using New Zealand marine snails with a superb fossil record (Crampton et al. 2003. Science 301: 358-360) and new DNA sequencing technology we plan to test rigorously the fundamental theory that cladogenesis is responsible for morphologically punctuated evolution.  fleshy fruits (
fleshy fruits (